UniBind: A map of direct TF-DNA interactions in the human genome
Description
We provide here the track hub that corresponds to the map of direct TF-DNA
interactions (aka TFBSs) stored in the UniBind
database.
UniBind is a comprehensive map of direct transcription factor (TF) - DNA
interactions in the human genome. These interactions were obtained by uniformly
processing ~2,000 public ChIP-seq data sets, from raw reads to high confidence
TF binding site predictions, using the ChIP-eat software. The
uniform processing, up to ChIP-seq peaks calling was performed by ReMap and the
entire collection of ChIP-seq peaks is also available in the ReMap database. ChIP-eat
used the MACS2 peak caller to identify ChIP-seq peaks on the hg38 version of
the human genome. An entropy-based algorithm was used to automatically
delineate an enrichment zone containing direct TF-DNA interactions, supported
by both strong computational evidence and strong experimental evidence. The
UniBind database hosts the complete set of TFBS predictions for each prediction
model, as well as the models themselves, the original ChIP-seq peaks, and
cis-regulatory modules derived from these direct TF-DNA interactions. All the
data is publicly available. For further details, please refer to the
associated publication: (DOI: https://doi.org/10.1093/nar/gky1210).
Individual BED files for specific TFs or datasets can be found and
downloaded on the UniBind website at http://unibind.uio.no.
Display Conventions and Configuration
-
Each transcription factor follow a specific RGB color.
- A set of TFBSs derived from a specific ChIP-seq experiment with a specific
TF binding profile from JASPAR is
defined with a name following the format
<GEO/ArrayExpress/ENCODE identifier>.<cell type/tissue>_<condition>.<TF name>.<JASPAR ID>.<JASPAR version>.<TF binding model>
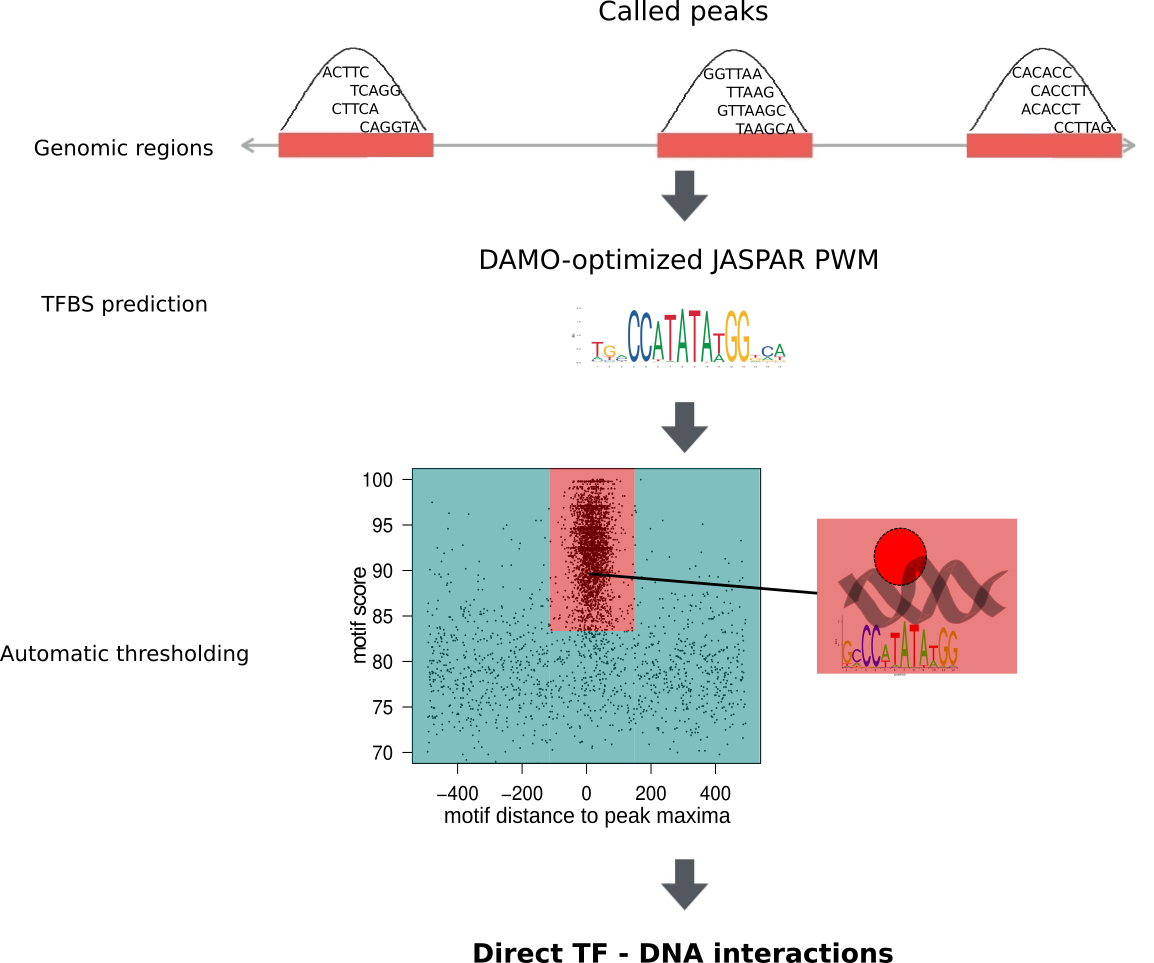
Methods
The entire collection of ChIP-seq data sets was uniformly processed in ReMap up
to ChIP-seq peak calling. The entire collection of ChIP-seq peaks is also
available in the ReMap database. These peaks served as input for the ChIP-eat
data processing pipeline. The complete pipeline is designed to uniformly
process ChIP-seq data sets, from raw reads to the identification of direct
TF-DNA binding events, and it was implemented in the ChIP-eat software with
source code freely available at https://bitbucket.org/CBGR/chip-eat/. Only the
ChIP-seq datasets for which a TF binding profile for the targeted TF was
available in JASPAR were used for TFBS predictions. The enrichment zone
containing high confidence direct TF-DNA interactions was automatically defined
for each data set using an entropy-based algorithm. The diagram below
illustrates the processing steps.
Data Availability
Individual BED files for specific TFs or datasets can be found and
downloaded on the UniBind website at http://unibind.uio.no.
Reference
If you use UniBind or ChIP-eat in your work, please cite:
M. Gheorghe, G.K. Sandve, A. Khan, J. Cheneby, B. Ballester, and A. Mathelier,
A map of direct TF-DNA interactions in the human genome.
Nucleic Acids Research (2019) gky1210 https://doi.org/10.1093/nar/gky1210.
Contact
If you have questions or comments, please write to:

