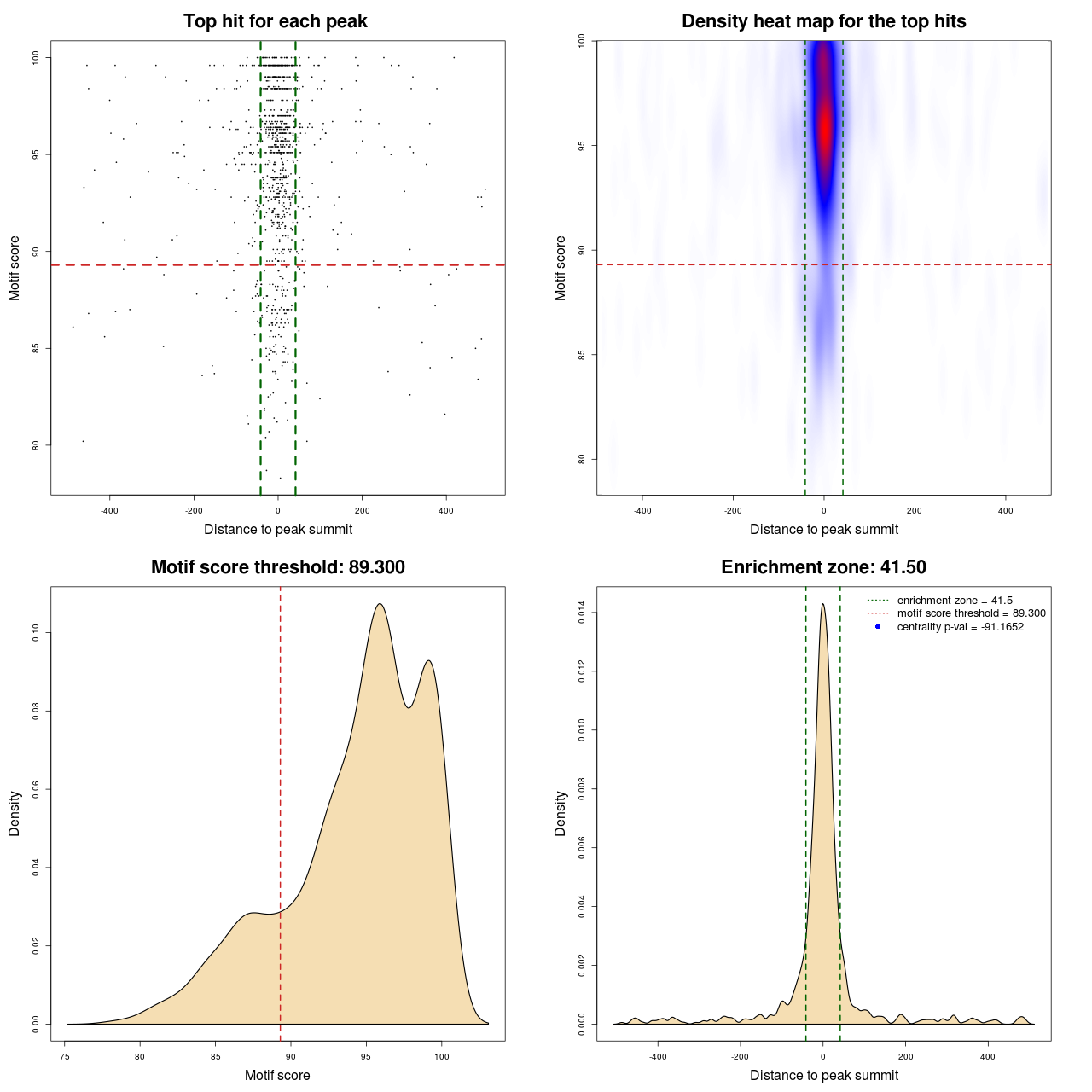
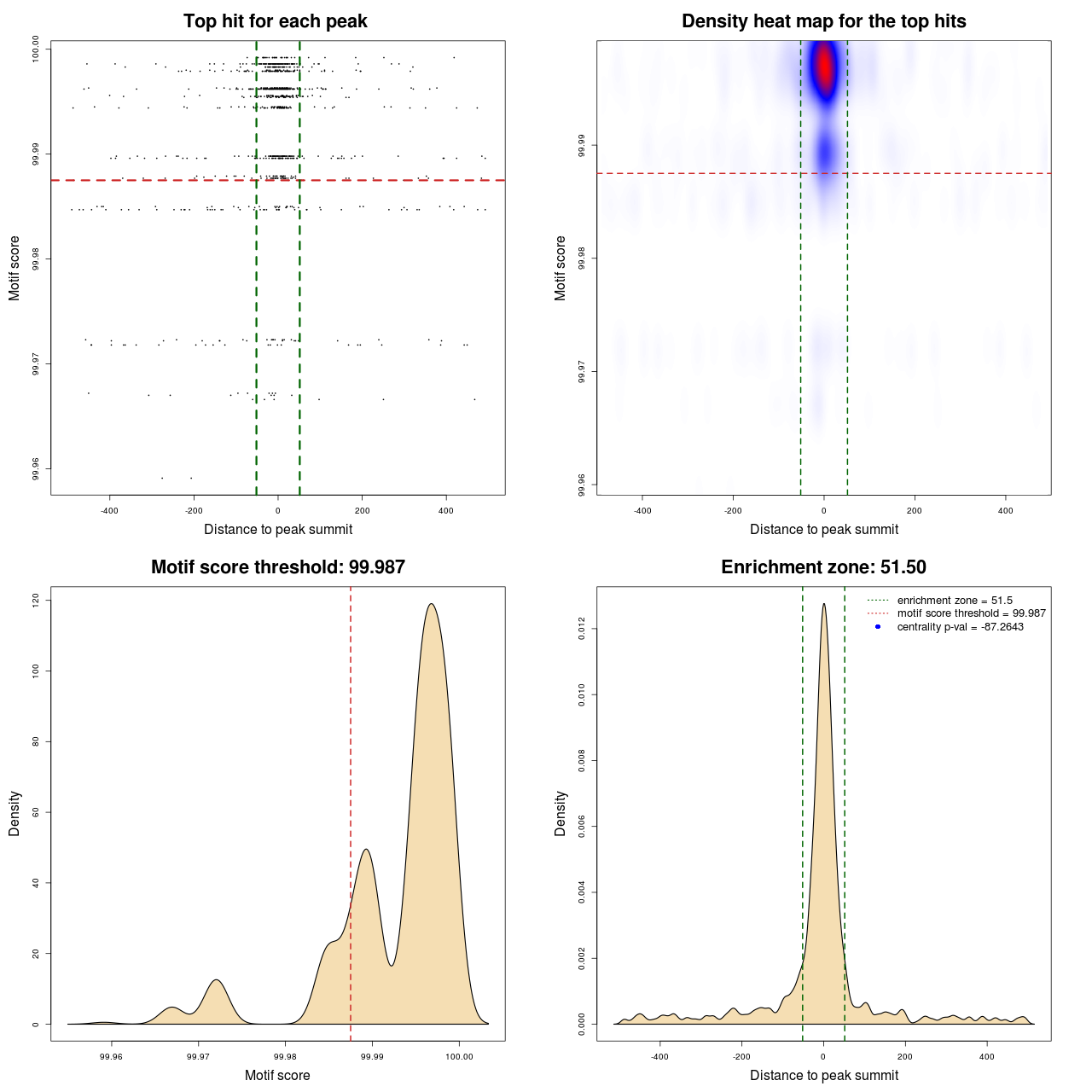
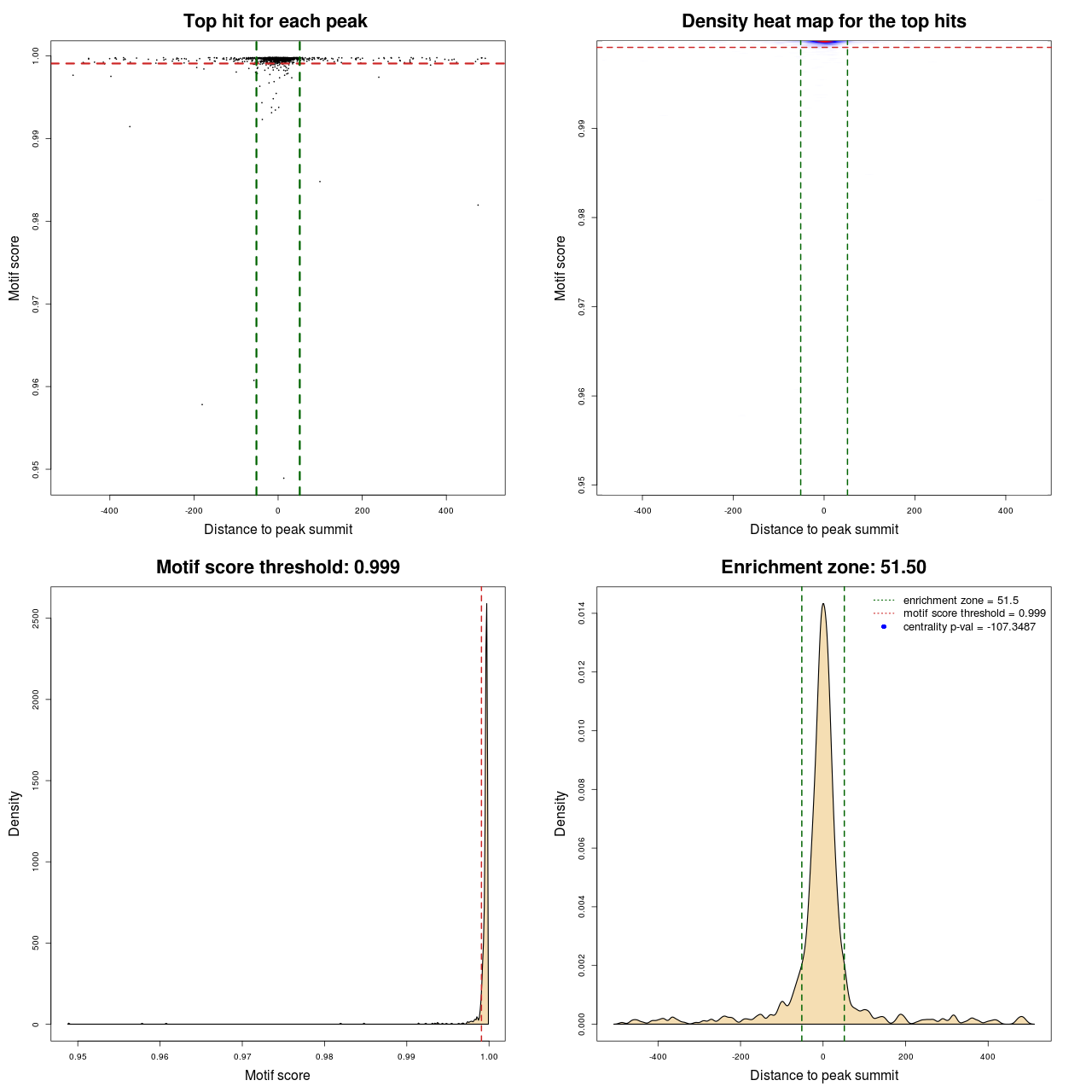
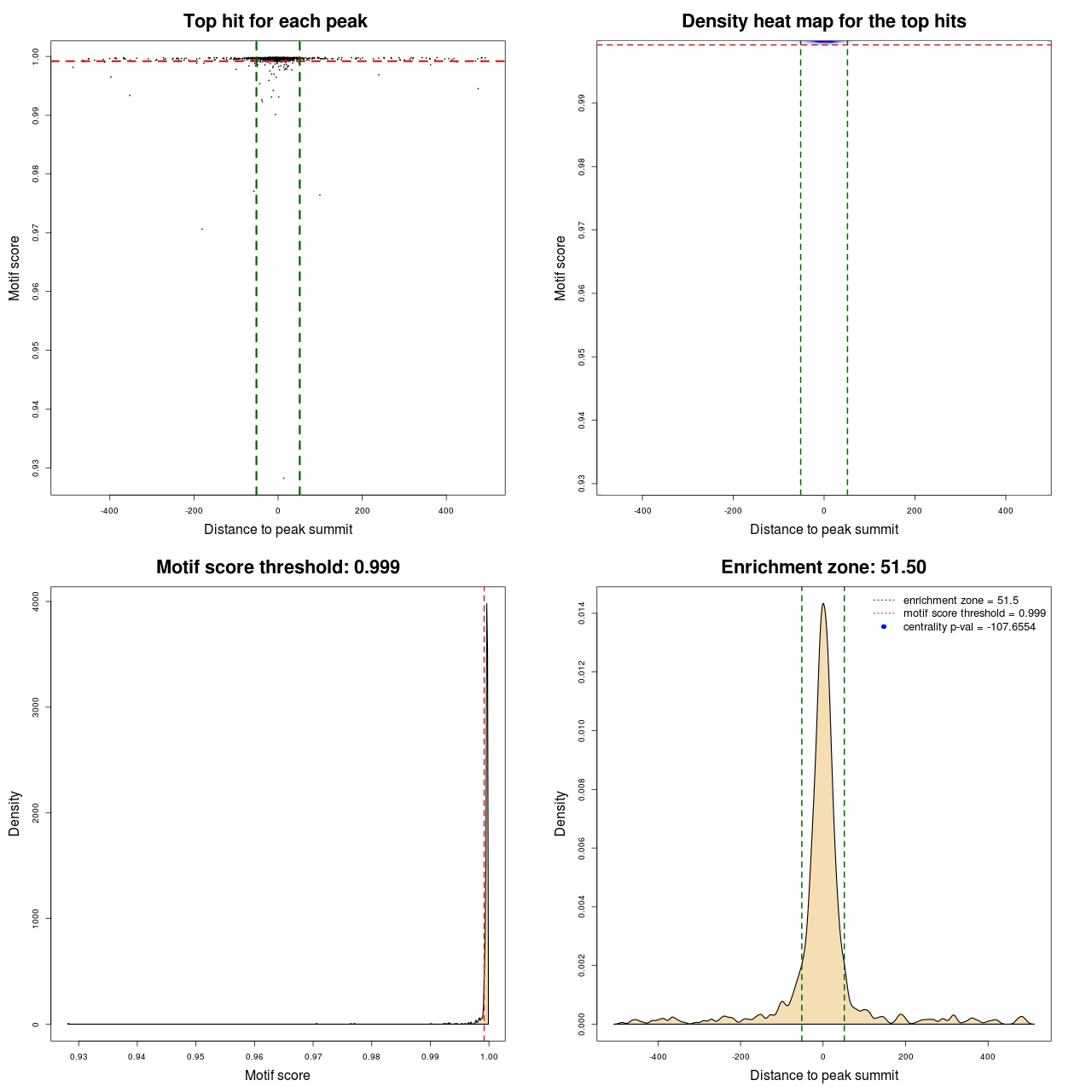
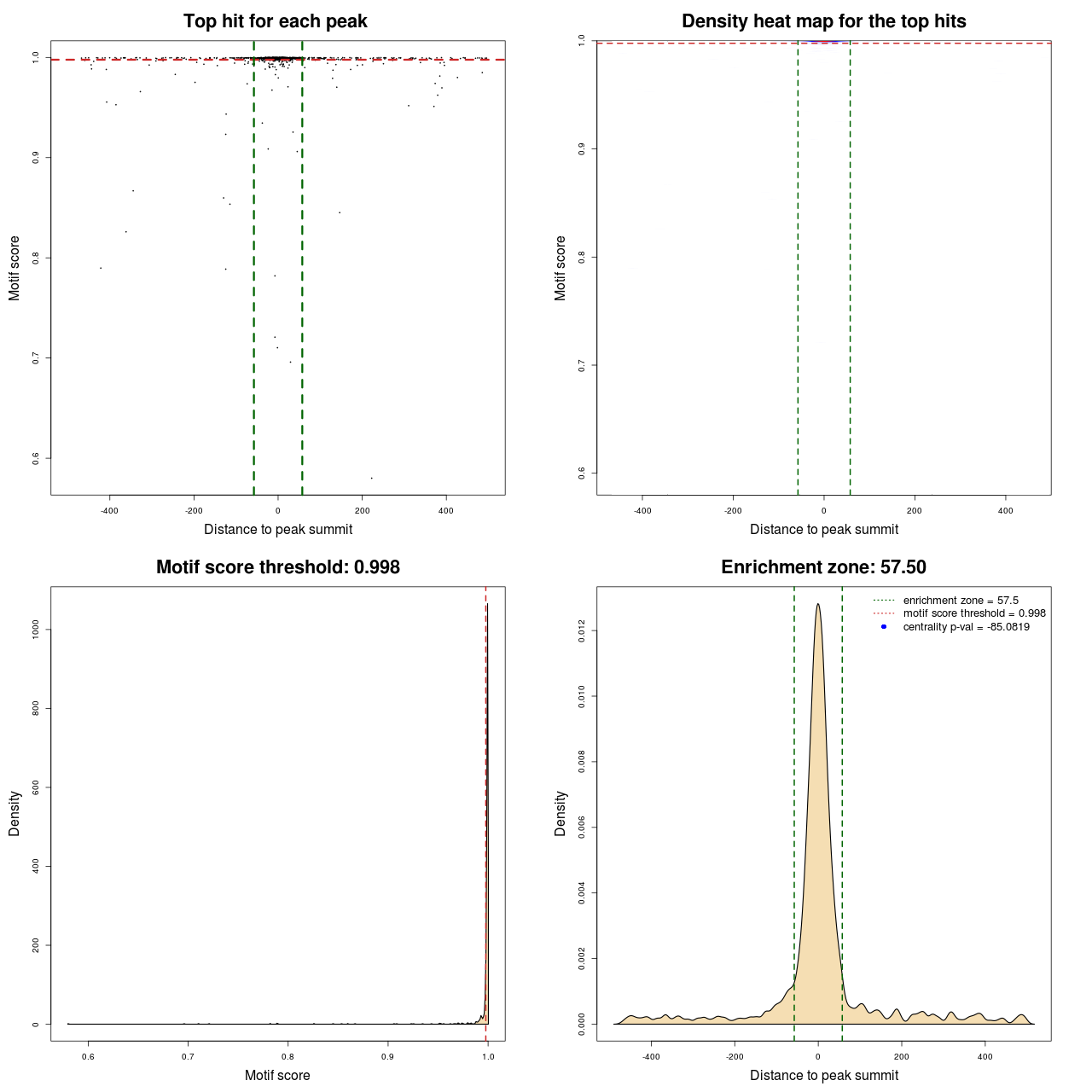
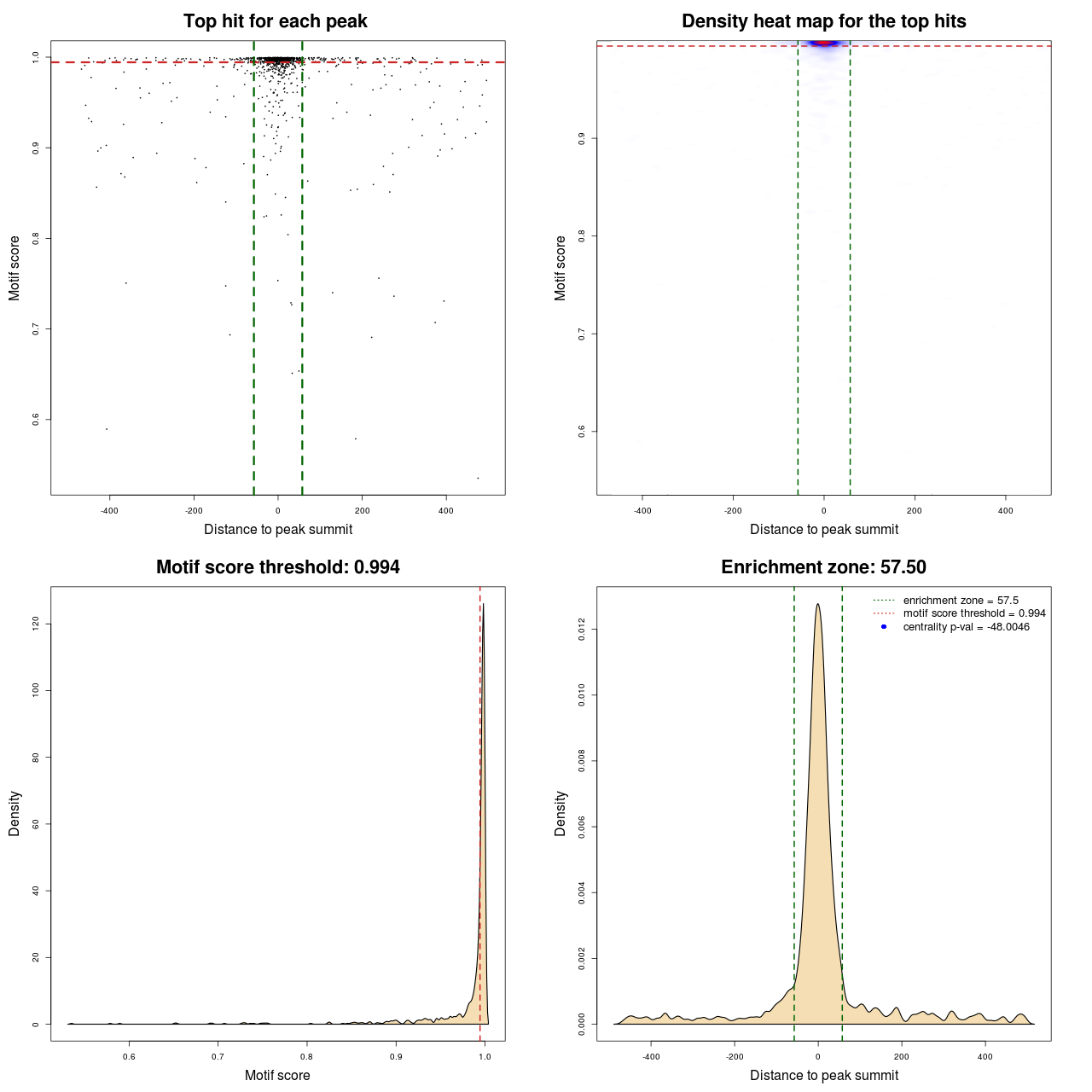
Statistical details
| Model | Peak caller | Model ID | Enrichment Zone | TFBSs | Score threshold | Centrality p-value (log10) |
|---|---|---|---|---|---|---|
| BEM | MACS | MA0095.2 | 103 | 708 | 99.9875 | -87.26428823451 |
| PWM | MACS | MA0095.2 | 83 | 674 | 89.3 | -91.165219395827 |
| DNAshaped | MACS | MA0095.2 | 103 | 759 | 0.99908 | -107.34871018718 |
| DNAshaped | MACS | MA0095.2 | 103 | 760 | 0.99918 | -107.65542850307 |
| DNAshaped | MACS | MA0095.2 | 115 | 704 | 0.99772 | -85.081864184958 |
| TFFM | MACS | MA0095.2 | 115 | 567 | 0.99433 | -48.004624395121 |
18 datasets found for the same TF (YY1) in other cell-types/tissues
| TF Name | Cell-line | Condition/Source | Collections | Species | Identifiers | JASPAR ID |
|---|---|---|---|---|---|---|
| YY1 | ESC | h1 | Permissive | Homo sapiens | ENCSR000BKD | MA0095.2 |
| YY1 | GM12891 | Permissive | Homo sapiens | ENCSR000BKJ | MA0095.2 | |
| YY1 | K562 | Permissive | Homo sapiens | ENCSR000BKU | MA0095.2 | |
| YY1 | GM12892 | Permissive | Homo sapiens | ENCSR000BLT | MA0095.2 | |
| YY1 | SKNSH | Permissive | Homo sapiens | ENCSR000BLZ | MA0095.2 | |
| YY1 | K562 | Permissive | Homo sapiens | ENCSR000BMH | MA0095.2 | |
| YY1 | GM12878 | Permissive | Homo sapiens | ENCSR000BNP | MA0095.2 | |
| YY1 | HEPG2 | Permissive | Homo sapiens | ENCSR000BNT | MA0095.2 | |
| YY1 | HCT166 | Permissive | Homo sapiens | ENCSR000BNX | MA0095.2 | |
| YY1 | A549 | Permissive | Homo sapiens | ENCSR000BPM | MA0095.2 | |
| YY1 | SKNSH | Permissive | Homo sapiens | ENCSR000BSM | MA0095.2 | |
| YY1 | ISHIKAWA | Permissive | Homo sapiens | ENCSR000BSY | MA0095.2 | |
| YY1 | GM12878 | Permissive | Homo sapiens | ENCSR000EUM | MA0095.2 | |
| YY1 | K562 | Permissive | Homo sapiens | ENCSR000EWF | MA0095.2 | |
| YY1 | NT2D1 | Permissive | Homo sapiens | ENCSR000EXG | MA0095.2 | |
| YY1 | LIVER | Permissive | Homo sapiens | ENCSR382MOM | MA0095.2 | |
| YY1 | LIVER | Permissive | Homo sapiens | ENCSR994YLZ | MA0095.2 | |
| YY1 | ESC | h1 | Permissive | Homo sapiens | GSE39096 | MA0095.2 |